
When investigating a crime, forensic scientists sometimes use DNA evidence to identify suspects. DNA contains more than identifying information, though, like clues to our genetic makeup. DNA separation is used to take a closer look at DNA strands, but traditional methods are time consuming. To speed up the DNA separation process, researchers from the Missouri University of Science and Technology turned to the COMSOL Multiphysics® software.
A Closer Look at Our Genetic Makeup
The molecular structure of DNA is complex: It’s a double-helix polymer made up of a long chain of nucleotides. Studying DNA is much easier by breaking up the sample into fragments of varying sizes.
The nucleotide, or base, pairs of DNA are guanine (G), adenine (A), thymine (T), and cytosine (C). Researchers try to make sense of the sequences of these genetic “letters” in areas like genome sequencing and medical diagnosis, for example, to locate genes and see how they work together within an organism. This work would not be easy to do without DNA separation — after all, the human genome has over 3 billion base pairs of DNA!
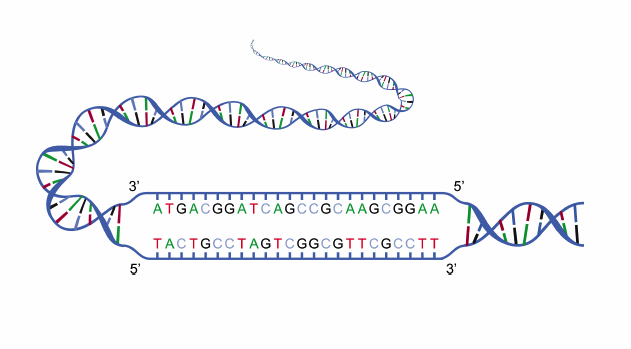
An illustration of DNA strands containing the base pair letters G, A, T, and C. Image in the public domain in the United States, via Wikimedia Commons.
Other examples of DNA analysis have entered the spotlight more recently. You may be familiar with mail-in DNA test kits that help you learn more about your ancestry. When a genetic testing company digitizes your DNA sample, it looks like a long strand of the nucleotide G, A, T, and C letters. These companies use algorithms to compare your piece of DNA from the genome against sets of reference data. Then, the algorithm determines how closely your DNA sample matches each reference set to see which ancestry groups you most likely belong to. The algorithm is only as good as its reference sets, so some ancestral groups may be underrepresented compared to others in the database.
In fields such as forensics, DNA profiling helps scientists compare samples of genetic material. Since it’s rare that two people have the same DNA pattern, forensic scientists can compare the patterns in slices of DNA molecules to reference databases, such as the Combined DNA Index System (CODIS) managed by the U.S. FBI. However, systems like CODIS are limited to the DNA profiles they contain. Investigators are starting to use ancestry databases like those mentioned above to expand their searches via a concept called familial DNA. For instance, in 2018, police investigating the case of the Golden State Killer ran the crime scene DNA against a genealogy site database and found a partial match to a distant relative. Ultimately, this helped them narrow down their search and identify the alleged suspect.
Investigating Fragmented Links in a Nucleotide Chain with DNA Separation
One common technique used to separate DNA molecules — primarily in forensics — is gel electrophoresis, which involves the migration of negatively charged nucleic acid molecules by means of a gel. When an electric current is applied, the smaller molecules move through the gel faster than the larger ones, thus the fragments are separated into bands based on size. To visualize this separation, radioactive dye is used.
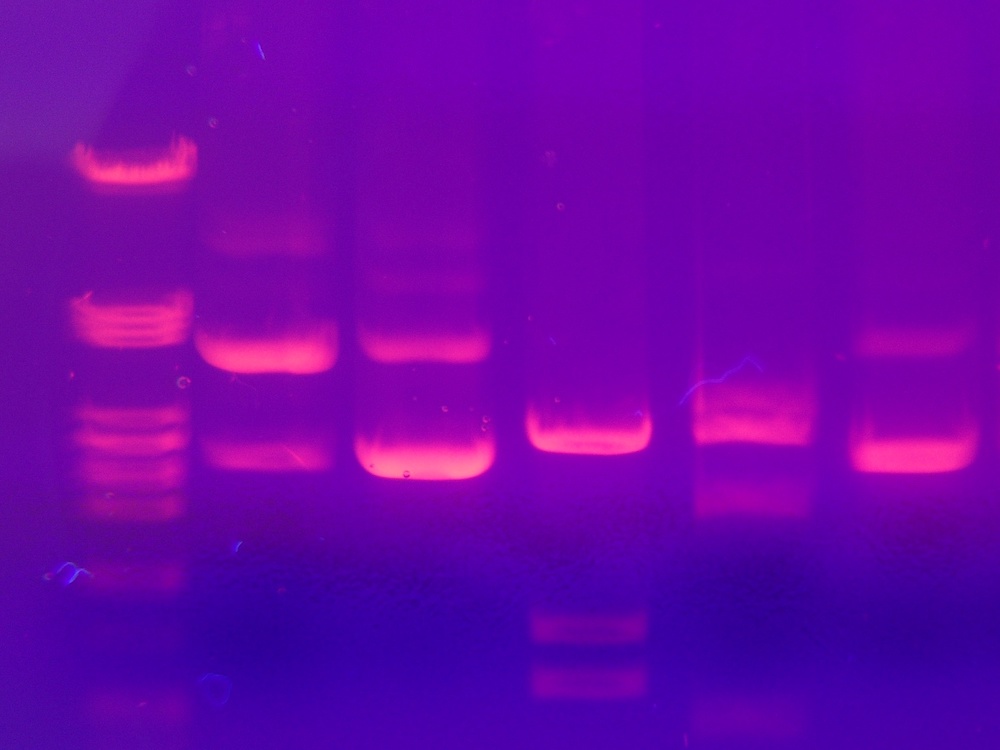
An example of gel electrophoresis results. Image by Mnolf — Own work. Licensed under CC BY-SA 3.0, via Wikimedia Commons.
There is another method that can separate long strands of DNA much more efficiently without the use of a gel or electric fields: entropic trapping. In this microchip-based system, entropic trap arrays (structured microchannels) of different heights are set up so that the narrow channel gap is much smaller than the gyration diameter of a DNA molecule. The molecules can be separated depending on the chain length. When negatively charged DNA molecules are driven through these channels by electrophoretic forces, the elution times are dependent on length. The longer the DNA molecule, the more likely it is to be drawn into the small channels, because the longer molecules occupy more surface area.

Schematic of an array of entropic traps, with a DNA molecule in a wide channel flowing into a narrow channel. Image courtesy Missouri University of Science and Technology.
Although entropic trapping is faster and more efficient than other separation methods, the design and fabrication of the devices needed takes a lot of time and can be costly, since it relies on trial-and-error. Since discovering the entropic trapping method, researchers have run computational studies to optimize designs and look into the separation mechanisms within these devices, but commercial software had yet to be used to simulate these entropic trap systems…until now.
Simulating Polymer Dynamics in an Entropic Trap System with COMSOL Multiphysics®
To find out whether they could save time with a commercially available simulation software, researchers from the Missouri University of Science and Technology set up their entropic trap system and polymer dynamics simulation using COMSOL Multiphysics® and compared their results to experimental data.
The research team, comprised of Joontaek Park, James Jones, Meyyamai Palaniappan, Saman Monjezi, and Behrouz Behdani, says that “DNA dynamics in microchannel simulation is challenging because two different simulations — field calculation in a complex microfluidics geometry and polymer molecule dynamics — must be combined.” Fortunately, they add, “COMSOL® can relatively easily handle the former simulation,” and that “COMSOL® can open a new page in the DNA or single-polymer molecule simulation area.”
Using the add-on Particle Tracing Module, the team performed Brownian dynamics simulations of the DNA chain. The chain was set up as a single-polymer, bead-chain model within a Newtonian fluid with the help of the CFD Module. As for the beads themselves, they were treated as Brownian particles to account for the random movements of the chain as it moves through the surrounding solvent.
To describe the spring force between each bead, they used another well-known model, the worm-like chain (WLC), which describes the behavior of semiflexible polymers. Alongside the WLC, the research team used the Lennard–Jones potential to keep the beads from penetrating each other. After setting up the entropic array geometry (shown below) so that Hs is much smaller than the gyration diameter of a typical DNA molecule, the researchers used the AC/DC Module to create the electric field of potential across the channel.
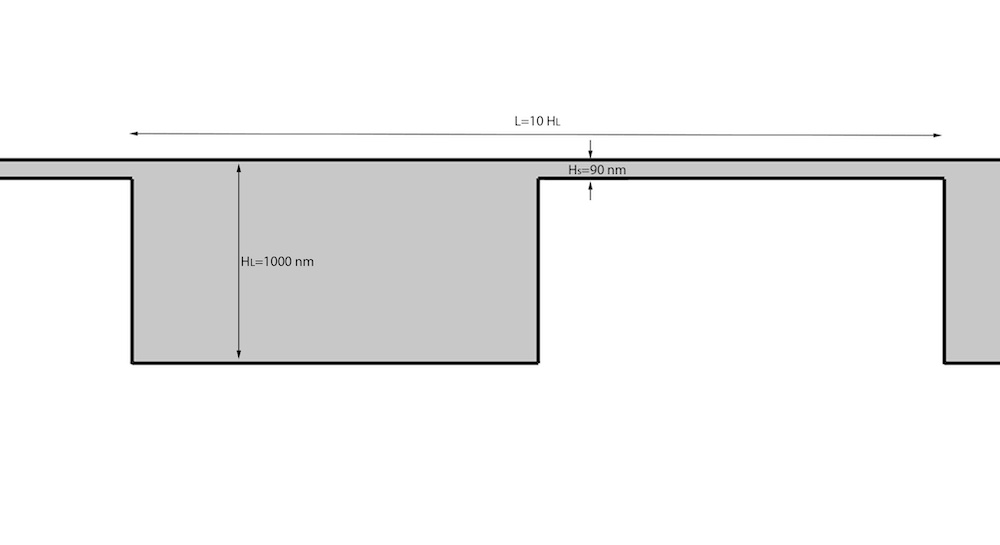
Schematic of the channel structures used in the simulation. Image courtesy Missouri University of Science and Technology.
Evaluating the Simulation Results
The research team calculated the nonuniform electric field using the finite element method. The electrical field direction can be seen here, with the arrows also indicating the direction of movement of the DNA molecules.
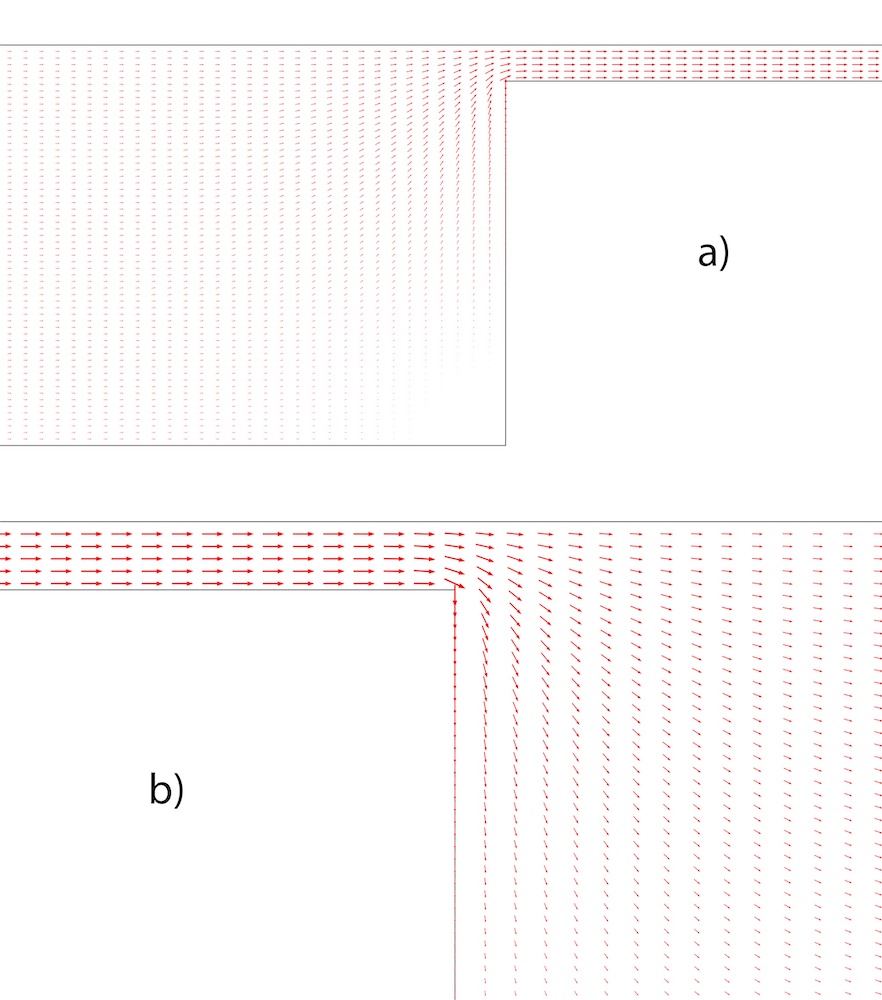
The electrical field flux vectors in the right corner (a) and the left corner (b) of a wide channel. Image courtesy Missouri University of Science and Technology.
Next, the researchers simulated the center-of-mass trajectory for the DNA molecules according to length, at Nb = 2, 4, and 16 bead lengths, as they periodically flowed into the constricted channel. The trajectories for each of the molecules traveling at the same distance are shown below. As expected from the electric field vector above, the molecules move faster in the narrow channels, and the longer the molecule (the more beads it has), the faster it moves. The shorter molecules, meanwhile, have a reduced velocity along their trajectory, and the distribution of DNA molecules indicates a more diffusive pattern, which reduces the overall velocity through the channels by moving them away from regions where the electric field is strongest.
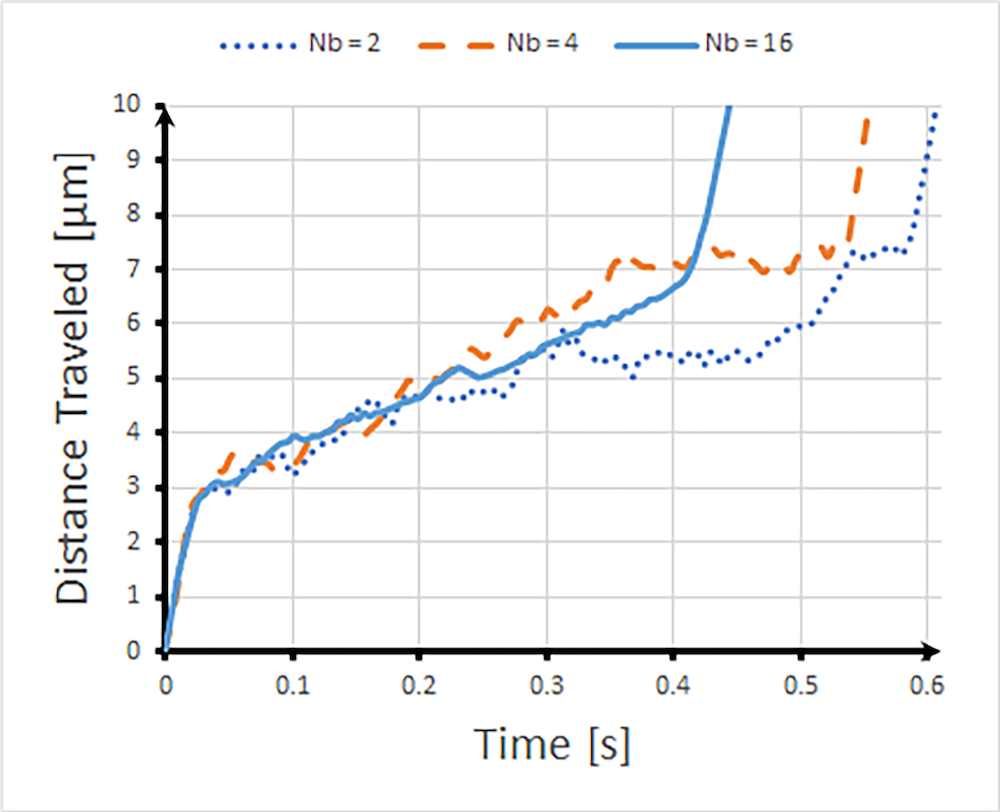
Center-of-mass trajectories of DNA molecules with Nb = 2, 4, and 16. Image courtesy Missouri University of Science and Technology.
This comparison can be seen in the animations below for the short bead length, Nb = 2, middle bead length Nb = 4, and the long bead length, Nb =16. As indicated by the units on the legend, the color in the animations shows the speed of the particles at any point in time. As expected, the larger the surface of a DNA molecule, the more likely it will be dragged into the smaller channel. (Note that the animations are ~10x slower than real time. If you prefer to slow the speed further, you can hover over the animation, then click the gear icon.)
Nb = 2 shorter DNA molecule flowing into and out of a wide channel in an entropic trap channel. Animation courtesy Missouri University of Science and Technology.
Nb = 4 middle-length DNA molecule flowing into and out of a wide channel in an entropic trap channel. Animation courtesy Missouri University of Science and Technology.
Nb = 16 longer molecule flowing into and out of a wide channel in an entropic trap channel. Animation courtesy Missouri University of Science and Technology.
The researchers were able to confirm that their simulation results were in good agreement with experimental data for the trajectory of DNA chains in an entropic trap, and these results show that the longer DNA chain, indeed, elutes faster than the shorter chain.
The use of COMSOL Multiphysics for polymer dynamics simulation has opened up possibilities for further research, as this was the first trial for this type of simulation using commercially available software. The team says that “COMSOL Multiphysics is a very popular and user-friendly simulation tool,” and further, that the extension of using the software for polymer dynamics will “enhance the related application and simulation studies.”
As for their own future research? The team adds that they could see pursuing the investigations on the inertia effect, the polymer configuration (branched polymer) effect, and DNA-carbon nanotube interaction.
Next Step
For more details about researchers’ work from the Missouri University of Science and Technology, click the button below:
Reference
- S. Monjezi, B. Behdani, M.B. Palaniappan, J.D. Jones, and J. Park, “Computational studies of DNA separations in micro-fabricated devices: Review of general approaches and recent applications”, Adv. in Chem. Eng. Sci., 7 (4), pp. 362–393, 2017.







Comments (0)