Buoyancy–driven μPCR for DNA Amplification
Application ID: 17227
Polymerase chain reaction (PCR) is one of the most effective methods in molecular biology, medical diagnostics, and biochemical engineering in amplifying a specific sequence of DNA. There has been a great interest in developing portable PCR-based lab-on-a-chip systems for point-of-care applications and one strategy that seems very promising is natural convection-based PCR.
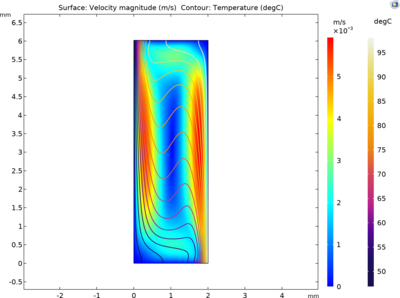
This model studies the spatio-temporal variation of the concentrations of single-stranded (ssDNA), double-stranded (dsDNA) and primer-annealed (aDNA) DNA components. The driving mechanism for the fluid motion is the temperature-induced density differences that results in buoyancy flow. This approach eliminates the need for an external driving mechanism and the difference between the temperatures is sufficient to circulate the PCR mixture and amplify DNA in a closed loop.
This model example illustrates applications of this type that would nominally be built using the following products:
however, additional products may be required to completely define and model it. Furthermore, this example may also be defined and modeled using components from the following product combinations:
The combination of COMSOL® products required to model your application depends on several factors and may include boundary conditions, material properties, physics interfaces, and part libraries. Particular functionality may be common to several products. To determine the right combination of products for your modeling needs, review the Specification Chart and make use of a free evaluation license. The COMSOL Sales and Support teams are available for answering any questions you may have regarding this.



